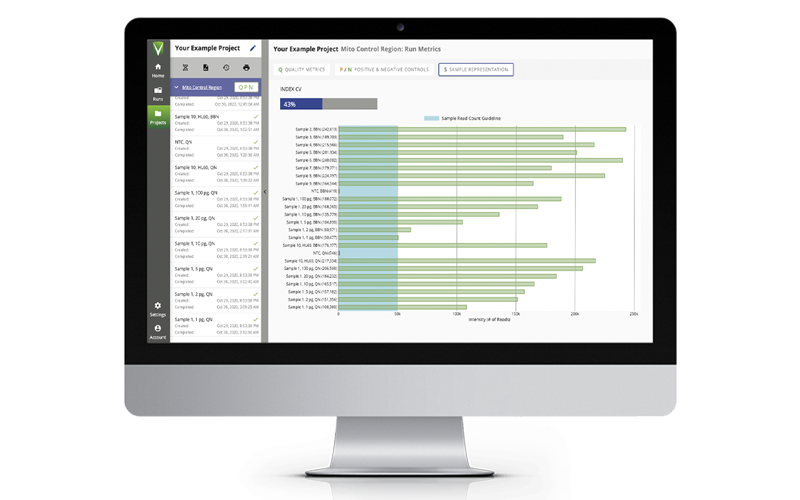
The ForenSeq mtDNA Control Region Kit offers convenient, efficient library prep for interrogating the control region of the mitochondrial genome (mtGenome) with minimal DNA input and optimum sensitivity. Leveraging proven ForenSeq chemistry, the kit produces a concise data set from the entire control region for direct upload to CODIS. Multiplexing capability boosts operational efficiency without increases to costs or time.
The ForenSeq mtDNA Control Region Kit is part of an integrated workflow that sequences libraries on the MiSeq FGx Sequencing System and analyzes data in Universal Analysis Software (UAS). Designed, developed, and manufactured as an end-to-end solution for high performance with forensic samples, extensive technical support backs the entire portfolio. Quality-controlled manufacturing ensures reproducibility with the convenience of all-in-one library prep kits.
Key Features
Specifications
| Target size | 1200 bp mtDNA control region |
| Sample type | extracted gDNA from a range of common forensic sample types |
| DNA input recommendation | 100 pg gDNA per sample |
| Recommended multiplexing capacity | 3–48 libraries per run |
| Number of primers | > 120 |
| Number of amplicons | 18 |
| Average amplicon size | 118 bp |
| Amplicon overlap | ≥ 3 bp |
| Total library prep time | 7 hours and 45 minutes |
| Hands-on time | 90 minutes |