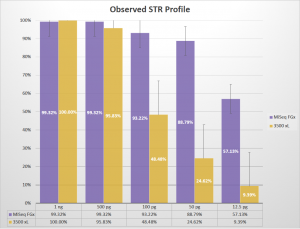
Observed profiles generated for STR targets by ForenSeq™ kit on MiSeq FGx™ vs. GlobalFiler™ kit on 3500 xL. (Figure 2 from WCU Poster – click image to enlarge.)
In a poster presented at the American Academy of Forensic Sciences 2018 Annual Scientific Meeting in Seattle, WA, a team at Western Carolina University (WCU) demonstrated the value of massively parallel sequencing (MPS) technology in analyzing forensically relevant samples. They performed a comparison of two commercially available kits, the ForenSeq™ DNA Signature Prep Kit using a MiSeq FGx™ sequencer versus the Applied Biosystems® GlobalFiler™ kit with a 3500 xL capillary electrophoresis (CE) instrument. The sensitivity experiments, using a dilution series ranging from 1ng down to 12.5pg of input DNA, showed that ForenSeq on the MiSeq FGx was capable of recovering a far greater number of STR alleles than the GlobalFiler kit using legacy CE instrumentation. See Figure 2 from the poster above. To view the entire poster, please click here.
The MiSeq FGx System successfully typed a significantly increased number of loci for all possible marker types, including global autosomal STRs, Y-STRs, X-STRs, identity SNPs, and phenotypic and biogeographical ancestry SNPs. However, even when only comparing the 20 common CODIS STR loci between methods, the MiSeq FGx showed substantially increased data recovery. See supplementary data figure from WCU below:
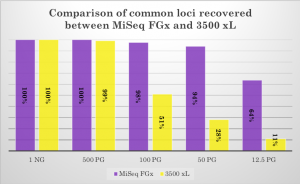
Comparison of data recovery for the 20 CODIS core STR loci common to both the ForenSeq™ kit run on MiSeq FGx™ and the GlobalFiler™ kit run on 3500 xL. (Supplementary data from WCU – click image to enlarge.)
At the lowest level of DNA input, 12.5pg of DNA, the ForenSeq kit was able to recover almost 6 times as many CODIS STR alleles as GlobalFiler, as well as a substantial number of additional markers.
The study, one of the first head-to-head comparisons of the legacy CE fragment analysis technology to the new MPS and ForenSeq library preparation technology, demonstrates some of the key advantages of the new approach to forensic genomics:
- By sequencing the individual markers, and removing the need for dye channels, the MPS approach enables significantly more markers to be included without performance degradation, thus increasing discrimination power.
- Fragment lengths are optimized for reliable amplification and analysis of all sample types as there is no need to fit markers into a dye channel.
- The MiSeq FGx™ Forensic Genomics System enables forensic examiners to generate substantially more probative data from challenging and/or limited biological evidence samples, for both “core” autosomal STR loci as well as additional markers to aid investigations.
For more detail on the experiments conducted at WCU, please click here to view the poster presentation.
Special thank you to Ashlee Volk for sharing her poster and supplementary data.
